New paper on phylogenomics of Heterochromatin Protein 1
22 Jun 2012, by Erick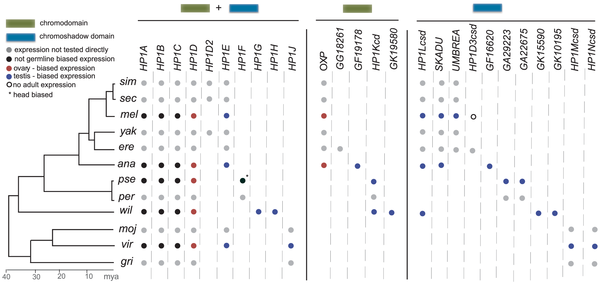 I don’t know the first thing about fly genetics.
Me explaining something about fly genetics is like someone
explaining the Star Wars trilogy who has never seen the movies in their entirety.
But, here goes.
Eukaryotic DNA is packed into in chromatin, which comes in two flavors: euchromatin and heterochromatin.
Heterochromatin is typically not transcriptionally active, and is chock-full of repetitive sequence.
However, it’s quite important for cell division.
The HP1 gene family is known to maintain heterochromatin, and knocking genes out from this family can have lethal consequences.
The conventional wisdom (e.g. Wikipedia)
is that HP1 genes have “chromo” and “chromoshadow” domains, linked by a “hinge” region.
I don’t know the first thing about fly genetics.
Me explaining something about fly genetics is like someone
explaining the Star Wars trilogy who has never seen the movies in their entirety.
But, here goes.
Eukaryotic DNA is packed into in chromatin, which comes in two flavors: euchromatin and heterochromatin.
Heterochromatin is typically not transcriptionally active, and is chock-full of repetitive sequence.
However, it’s quite important for cell division.
The HP1 gene family is known to maintain heterochromatin, and knocking genes out from this family can have lethal consequences.
The conventional wisdom (e.g. Wikipedia)
is that HP1 genes have “chromo” and “chromoshadow” domains, linked by a “hinge” region.
Well, it turns out that when Mia Levine, a postdoc in Harmit Malik’s lab, searched fly genomes to find more members of the HP1 gene family, she found something really strange. There were some HP1s that didn’t have chromo domains, and some that didn’t have chromoshadow domains. These aren’t just degraded genes: they are quite conserved at the codon level and transcriptionally active. When we helped them build trees on these genes, a picture emerged with many paralogous HP1’s. Strangely enough, though, there are diagonals in the gene presence/absence table, showing a “revolving door” of HP1 paralogs (see image). It’s safe to say we don’t completely understand what’s going on, but Mia is working hard on characterizing the function and expression localization of these genes. Here’s the paper describing what we found. More fun with Connor and the Malik group!