Sometimes projects take years to bear fruit.
As I described previously, Aaron Darling and I have been thinking for a good while about using Sequential Monte Carlo (SMC) for online Bayesian phylogenetic inference, in which an existing posterior on trees can be updated with additional sequences. In fact, we had a good enough proof-of-concept implementation in 2013 to give a talk at the Evolution meeting. The other talk from my group that year was about a surrogate function for likelihoods parameterized by a single branch length, which we called “lcfit”. These two projects have just recently resulted in intertwined submitted papers.
The SMC implementation, which we called sts for Sequential Tree Sampler, lay dormant for a while after Connor McCoy left the group despite a few efforts to rekindle it.
One of the things that kept us from wrapping it up was problems with particle degeneracy, which is as follows.
I think of SMC as a probabilistically correct version of evolutionary computation, in which trees get to reproduce if they have a high posterior.
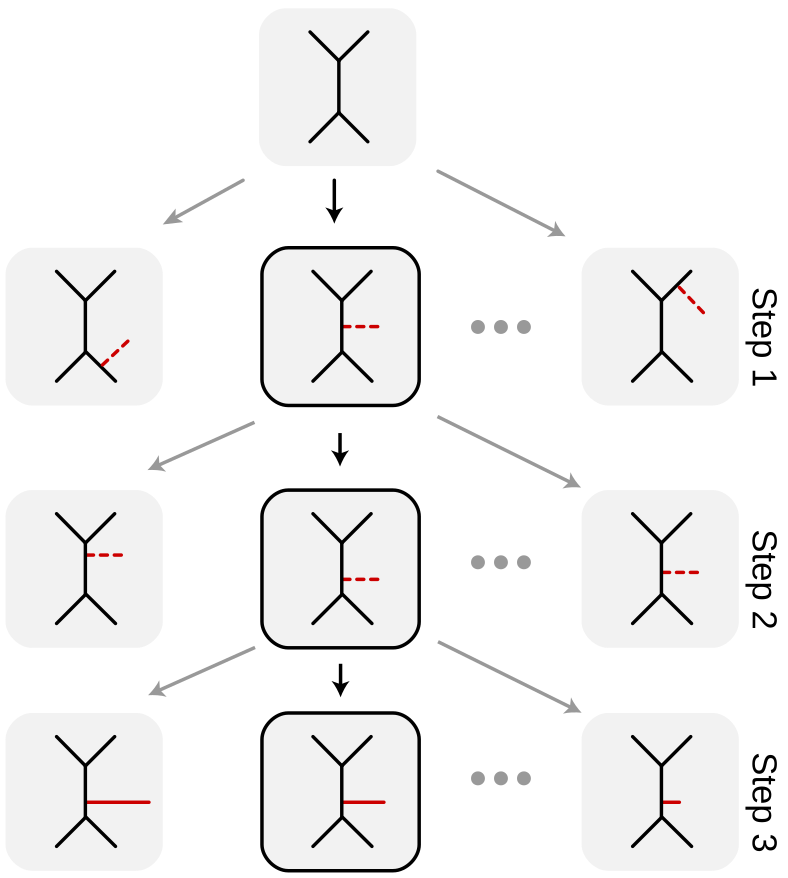
Every time we have a new sequence, we attach it to our existing population of trees via some proposal distribution suggesting where to add it.
Particle degeneracy, then, means that you obtain a low diversity population due to a few trees “taking over” the population because they are significantly better than the rest.
In working with sts, we found that we could mitigate the effect of particle degeneracy by developing “smart” proposals that use the data and corresponding likelihood function to decide where to try next.
This is in contrast to previous work by Bouchard-Côté et al 2012, which shows that for a different formulation of SMC based on subtree merging one does better with simple proposals (compare Teh et al. 2008).
Our proposals have to decide to what edge to attach, where along the edge to attach, and how long of a branch length to use for the attachment (see picture above).
Although the group put in hard work, there was a lot more needed to put all this together and actually get a working sampler. Luckily, Aaron recruited a super sharp and motivated postdoc in Mathieu Fourment. Mathieu tried a lot of variants of proposal distributions, showed that “heated” proposal distributions were effective for picking attachment branches, and added a parsimony-based proposal. He also showed that a high effective sample size is necessary but not sufficient to have a good SMC posterior sample, and that we can develop a time-competitive sampler compared to running MrBayes again and again. That work is now up on bioRxiv.
Smart proposals need to be designed carefully. One of the bottlenecks was proposing the new pendant branch length, and for that the lcfit surrogate function (the subject of Connor’s talk at Evolution 2013) worked well. This spurred us to finish and write up the lcfit work, despite the fact that Aberer et al. 2015 wrote a paper in which they describe how to use standard probability distribution functions as surrogate functions for branch length proposals. This took a bit of wind out of our sails, but our purpose-built lcfit surrogate still has some interesting advantages over common functions, such as that has the right “shape” for likelihood curves, even when branch lengths become long. We were surprised to find that it does well for complex settings with heterogeneous models. On a practical level, it’s implemented as a stand-alone C library so it can be easily incorporated into other programs, in contrast to the work of Aberer and co, which is tightly integrated into ExaBayes. In the end we have turned it into a short paper with a long appendix which is now up on arXiv. Brian Claywell is the hero of this lcfit work– he was persistent in developing creative strategies to fit the surrogate function in a variety of settings.
There is a lot to be done for online Bayesian phylogenetics still. We don’t even try to sample model parameters, and haven’t tried making BEASTly rooted trees. There are many opportunities for optimization, some of which are described in the paper. For the parallelism nerds out there, I also note the existence of the particle cascade.
I certainly hope that this contribution guides future developers towards useful online Bayesian phylogenetics samplers: it would be great to be able to keep trees up to date as the genomes keep rolling in from projects such as ZIBRA. If you are interested in more details, get in touch!